Updated April 10, 2023

Introduction to OpenCV watershed
There are several algorithms for the purpose of segmentation and one such algorithm is the watershed algorithm using which the touching or overlapping objects in a given image can be extracted and for the functioning of the algorithm, user-defined markers are necessary which can be defined manually by point and click or user-defined methods can be defined automatically using thresholding methods or morphological operations and proper placement of these markers is very much essential for accurate segmentation by watershed algorithm and watershed algorithm can be implemented using a function called watershed() function in OpenCV and the watershed() function returns a matrix of labels.
Syntax to define watershed() function in OpenCV:
watershed(-Euclideandistance, markers, mask)Where,
- Euclideandistance is the Euclidean distance to the closest zero for each of the foreground pixels in the given image that is to be segmented.
- markers are the user-defined markers defined manually by point and click or defined automatically using thresholding methods or morphological operations.
- the mask is the mask operation to be performed on the resulting image and it is optional.
Working of watershed() Function in OpenCV
- Computing the Euclidean Distance Transform or EDT is the first step in image segmentation using a watershed algorithm.
- The function distance_transform_edt is used to compute the Euclidean distance transform and returns the distance map.
- Then the local max is calculated using peak_local_max function.
- Then a connected component analysis is performed on the local peaks using 8 connectivity.
- Then the watershed function is applied by passing the negative of Euclidean distance and markers as the parameters.
- The watershed function returns a NumPy array which is a matrix of labels having the same dimensions as the input image.
- The unique objects in the image can be extracted by iterating through the unique label values obtained by using the watershed function.
Examples of OpenCV watershed
Given below are the examples of OpenCV watershed:
Example #1
OpenCV program in python to implement a watershed algorithm to perform segmentation of the given image by making use of watershed() function.
Code:
#importing all the required packages
from skimage.feature import peak_local_max
from skimage.morphology import watershed
from scipy import ndimage
import numpy as np
import argparse
import imutils
import cv2
#making use of argument parser to parse the input image that is to be segmented
imageparse = argparse.ArgumentParser()
imageparse.add_argument("-i", "--image", required=True, help="path to input image")
args = vars(imageparse.parse_args())
#performing thresholding and morphological operations after the input image is loaded
imageread = cv2.imread(args["image"])
op1 = cv2.pyrMeanShiftFiltering(imageread, 21, 51)
cv2.imshow("Input_image", imageread)
imagegray = cv2.cvtColor(op1, cv2.COLOR_BGR2GRAY)
imagethreshold = cv2.threshold(imagegray, 0, 255, cv2.THRESH_BINARY | cv2.THRESH_OTSU)[1]
cv2.imshow("Thresholded_image", imagethreshold)
#computing the Euclidean distance using distance_transform_edt function and then calculating the local max using peak_local_max function
EDT = ndimage.distance_transform_edt(imagethreshold)
localMax = peak_local_max(EDT, indices=False, min_distance=20, labels=imagethreshold)
#component connected analysis is performed on the local peaks using 8 connectivity and then applying the watershed function to display the number of segments returned by it
imagemarkers = ndimage.label(localMax, structure=np.ones((3, 3)))[0]
resultinglabels = watershed(-EDT, imagemarkers, mask=imagethreshold)
#iterating through the unique labels obtained by using watershed function
for eachlabel in np.unique(resultinglabels):
if eachlabel == 0:
continue
objectmask = np.zeros(imagegray.shape, dtype="uint8")
objectmask[resultinglabels == eachlabel] = 255
objects = cv2.findContours(objectmask.copy(), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
objects = imutils.grab_contours(objects)
result = max(objects, key=cv2.contourArea)
#drawing a circle around each unique object in the image
((x, y), r) = cv2.minEnclosingCircle(result)
cv2.circle(imageread, (int(x), int(y)), int(r), (0, 255, 0), 2)
cv2.putText(imageread, "#{}".format(eachlabel), (int(x) - 10, int(y)), cv2.FONT_HERSHEY_SIMPLEX, 0.6, (0, 0, 255), 2)
# displaying the resulting image as the output on the screen
cv2.imshow("Output_image", imageread)
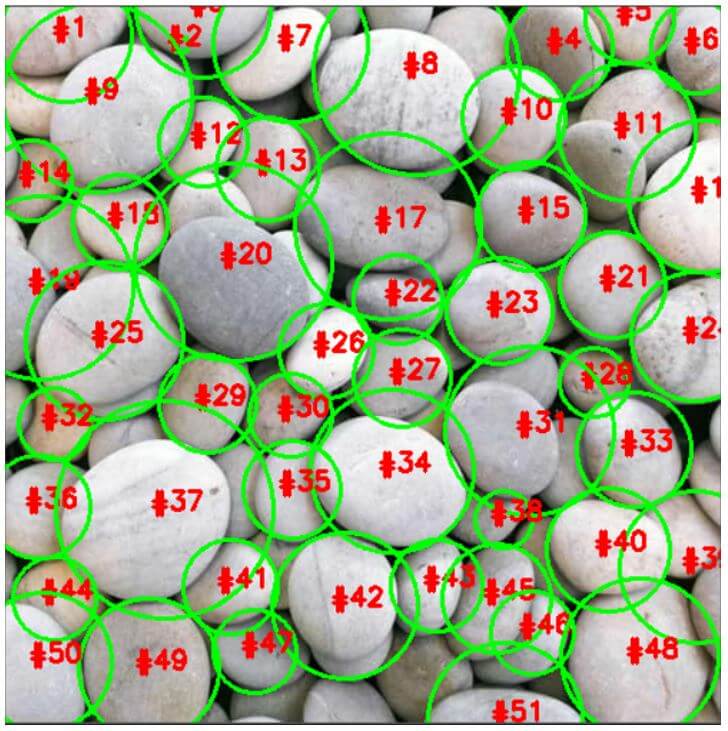
cv2.waitKey(0)Output:
In the above program, we are importing the required modules. Then we are making use of argument parser to parse the input image that is to be segmented. Then we are performing thresholding and morphological operations after the input image is loaded. Then we are computing the Euclidean distance and then calculating the local max. Then component connected analysis is performed on the local peaks using 8 connectivity and then applying the watershed function to display the number of segments returned by it. Then we are iterating through the unique labels obtained by using watershed function. Then we are drawing a circle around each unique object in the image and then displaying the resulting image as the output on the screen.
Example #2
OpenCV program in python to implement watershed algorithm to perform segmentation of the given image by making use of watershed() function.
Code:
#importing all the required packages
from skimage.feature import peak_local_max
from skimage.morphology import watershed
from scipy import ndimage
import numpy as np
import argparse
import imutils
import cv2
#making use of argument parser to parse the input image that is to be segmented
imageparse = argparse.ArgumentParser()
imageparse.add_argument("-i", "--image", required=True, help="path to input image")
args = vars(imageparse.parse_args())
#performing thresholding and morphological operations after the input image is loaded
imageread = cv2.imread(args["image"])
op1 = cv2.pyrMeanShiftFiltering(imageread, 21, 51)
cv2.imshow("Input_image", imageread)
imagegray = cv2.cvtColor(op1, cv2.COLOR_BGR2GRAY)
imagethreshold = cv2.threshold(imagegray, 0, 255, cv2.THRESH_BINARY | cv2.THRESH_OTSU)[1]
cv2.imshow("Thresholded_image", imagethreshold)
#computing the Euclidean distance using distance_transform_edt function and then calculating the local max using peak_local_max function
EDT = ndimage.distance_transform_edt(imagethreshold)
localMax = peak_local_max(EDT, indices=False, min_distance=20, labels=imagethreshold)
#component connected analysis is performed on the local peaks using 8 connectivity and then applying the watershed function to display the number of segments returned by it
imagemarkers = ndimage.label(localMax, structure=np.ones((3, 3)))[0]
resultinglabels = watershed(-EDT, imagemarkers, mask=imagethreshold)
#iterating through the unique labels obtained by using watershed function
for eachlabel in np.unique(resultinglabels):
if eachlabel == 0:
continue
objectmask = np.zeros(imagegray.shape, dtype="uint8")
objectmask[resultinglabels == eachlabel] = 255
objects = cv2.findContours(objectmask.copy(), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
objects = imutils.grab_contours(objects)
result = max(objects, key=cv2.contourArea)
#drawing a circle around each unique object in the image
((x, y), r) = cv2.minEnclosingCircle(result)
cv2.circle(imageread, (int(x), int(y)), int(r), (0, 255, 0), 2)
cv2.putText(imageread, "#{}".format(eachlabel), (int(x) - 10, int(y)), cv2.FONT_HERSHEY_SIMPLEX, 0.6, (0, 0, 255), 2)
# displaying the resulting image as the output on the screen
cv2.imshow("Output_image", imageread)
cv2.waitKey(0)Output:
In the above program, we are importing the required modules. Then we are making use of argument parser to parse the input image that is to be segmented. Then we are performing thresholding and morphological operations after the input image is loaded. Then we are computing the Euclidean distance and then calculating the local max. Then component connected analysis is performed on the local peaks using 8 connectivity and then applying the watershed function to display the number of segments returned by it. Then we are iterating through the unique labels obtained by using watershed function. Then we are drawing a circle around each unique object in the image and then displaying the resulting image as the output on the screen.
Conclusion
In this article, we have seen the concept of implementation of watershed algorithm using watershed() function.
Recommended Articles
We hope that this EDUCBA information on “OpenCV watershed” was beneficial to you. You can view EDUCBA’s recommended articles for more information.